GPerturb - Gaussian process modelling of single-cell perturbation data
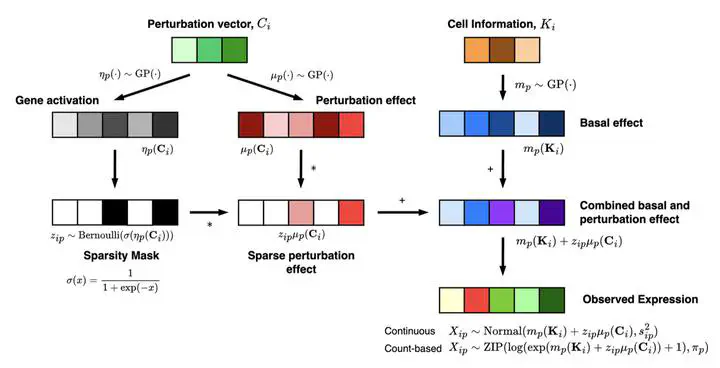
In this work, we introduce GPerturb, a new computational framework that helps make sense of high-throughput single-cell experiments in which genes are perturbed (for example via CRISPR) and the outcomes captured at single-cell resolution. The model uses Gaussian process regression within a hierarchical, additive structure to separate baseline (unperturbed) gene expression from the effect of perturbations, and to provide interpretable, uncertainty-aware estimates of how each gene responds.
Because single-cell perturbation data are often sparse, high‐dimensional and complex, GPerturb addresses these challenges by:
- modelling both continuous and count‐based expression data (via normal or zero‐inflated Poisson likelihoods) in a unified way.
- providing gene‐level effect estimates (rather than black‐box latent embeddings), making results more interpretable for biological follow‐up.
- demonstrating that its performance is competitive with state‐of‐the‐art deep-learning approaches across multiple datasets (single‐gene, multi‐gene, dosage perturbations) while retaining interpretability and flexibility.
In short: this work presents a scalable and interpretable model for analysing complex single-cell perturbation datasets, offering new opportunities to uncover how genes respond to perturbations (including multi-gene and dosage effects) and to support downstream biological discovery.
Investigator: