The Automatic BioData Scientist
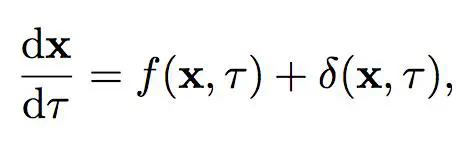
Mathematical models of disease evolution seek to specify the complex mechanistic relationships between many measurable quantities over space and time. However, as our capability to generate experimental data improves, our ability to conceive of appropriate mathematical descriptions to describe biological phenomena has become a bottleneck.
The goal of machine learning is to “learn” complex dependencies automatically within data sets. Neural networks (NNs) have been at the forefront of recent advances by offering a means of parametrising complex functional mapping between data and its representations.
Novel computational machinery have enabled NN approaches to scale to the analysis of unprecedently large datasets and provide versatility over standard modelling approaches. However, despite their successes in a range of applications, in biomedical research, NNs are often derided for their “black box” discoveries, lack of interpretability and the need for unrealistic quantities of training data. Such criticisms often overlook the fact that default NN structures express no explicit assumptions about the problems on which they are applied and they are often used as generic data mining devices for discovery purposes.
Nonetheless, there is a clear opportunity for the development of novel NN approaches that combine scalability and versatility with the capability of learning the physically realistic constraints that are embedded in hand-crafted mathematical models.
This project seeks to investigate the methodological foundations for what could form an Automated BioData Scientist (AutoBioDataSci) platform. We would like to develop learning algorithms that can capture biological laws and encapsulate them in such a way that such knowledge can be transfer to new problems. This major project involves inter-linking representation learning theory with transfer and meta-learning as well as having its roots embedded in the biological scientific discovery process.
Researchers: