Cancer
Congratulations to Hanwen Xing on having his paper “GPerturb: Gaussian process modelling of single-cell perturbation data” accepted in Nature Communications:
“Single-cell RNA sequencing and CRISPR screening enable high-throughput analysis of genetic perturbations at single-cell resolution. Understanding combinatorial perturbation effects is essential but challenging due to data sparsity and complex biological mechanisms. We present GPerturb, a Gaussian process-based sparse perturbation regression model designed to estimate gene-level perturbation effects. GPerturb employs an additive structure to separate signal from noise and captures sparse, interpretable effects from both discrete and continuous responses. It also provides uncertainty estimates for the presence and strength of perturbation effects on individual genes. We demonstrate the use GPerturb on both simulated and real-world datasets, characterising its competitive performance with current state-of-the-art methods. Furthermore, the model reveals meaningful gene-perturbation interactions and identifies effects consistent with known biology. GPerturb offers a novel approach for uncovering complex dependencies between gene expression and perturbations and advancing our understanding of gene regulation at the single-cell level.”
Congratulations to Charles Gadd on having his paper accepted at MLCB 2024.
Changes in the number of copies of certain parts of the genome, known as copy number alterations (CNAs), due to somatic mutation processes are a hallmark of many cancers. This genomic complexity is known to be associated with poorer outcomes for patients but describing its contribution in detail has been difficult. Copy number alterations can affect large regions spanning whole chromosomes or the entire genome itself but can also be localised to only small segments of the genome and no methods exist that allow this multi-scale nature to be quantified. In this paper, we address this using Wave-LSTM, a signal decomposition approach designed to capture the multi-scale structure of complex whole genome copy number profiles. Using wavelet-based source separation in combination with deep learning-based attention mechanisms. We show that Wave-LSTM can be used to derive multi-scale representations from copy number profiles which can be used to decipher sub-clonal structures from single-cell copy number data and to improve survival prediction performance from patient tumour profiles.
Congratulations to Kaspar Martens on having his paper accepted in Bioinformatics.
Cell type identification plays an important role in the analysis and interpretation of single-cell data and can be carried out via supervised or unsupervised clustering approaches. Supervised methods are best suited where we can list all cell types and their respective marker genes a priori, while unsupervised clustering algorithms look for groups of cells with similar expression properties. This property permits the identification of both known and unknown cell populations, making unsupervised methods suitable for discovery. Success is dependent on the relative strength of the expression signature of each group as well as the number of cells. Rare cell types therefore present a particular challenge that is magnified when they are defined by differentially expressing a small number of genes.
Congratulations to Joel Nulsen on having his paper accepted in BMC Bioinformatics.
Genomic insights in settings where tumour sample sizes are limited to just hundreds or even tens of cells hold great clinical potential, but also present significant technical challenges. We previously developed the DigiPico sequencing platform to accurately identify somatic mutations from such samples.
Congratulations to Kaspar Martens on having his paper accepted at MLCB 2023.
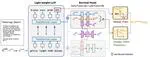
Generative models for multimodal data permit the identification of latent factors that may be associated with important determinants of observed data heterogeneity. Common or shared factors could be important for explaining variation across modalities whereas other factors may be private and important only for the explanation of a single modality. Multimodal Variational Autoencoders, such as MVAE and MMVAE, are a natural choice for inferring those underlying latent factors and separating shared variation from private. In this work, we investigate their capability to reliably perform this disentanglement. In particular, wehighlight a challenging problem setting where modality-specific variation dominates the shared signal. Taking a cross-modal prediction perspective, we demonstrate limitations of existing models, and propose a modification how to make them more robust to modalityspecific variation. Our findings are supported by experiments on synthetic as well as various real-world multi-omics data sets. The paper is available on PMLR.
Our latest paper, based on the thesis work of former DPhil student Zhiyuan, has been published in Genome Biology. CIDER: an interpretable meta-clustering framework for single-cell RNA-seq data integration and evaluation describes a meta-clustering workflow based on inter-group similarity measures. We demonstrate that CIDER outperforms other scRNA-Seq clustering methods and integration approaches in both simulated and real datasets. Moreover, we show that CIDER can be used to assess the biological correctness of integration in real datasets, while it does not require the existence of prior cellular annotations.
Kaspar Martens will present his latest work at the NeurIPS Workshop “Learning Meaningful Representations of Life” Rarity: Discovering rare cell populations from single-cell imaging data. The work arises from his Turing-Crick Biomedical Award which supports a collaboration between the Alan Turing Institute and the Ciccarelli Group at Kings College London and the Francis Crick Institute.
The Columbia Hospital For Women Research Foundation have awarded Christopher Yau the prize for most impactful paper in 2020 in the field of obstetrical and gynecologic and breast disease. The prize consisted of a $5,000 to a charity of Chris and Ahmed’s choosing and they selected Ovarian Cancer Action who co-funded the original work.