Multi-Omics Network inference
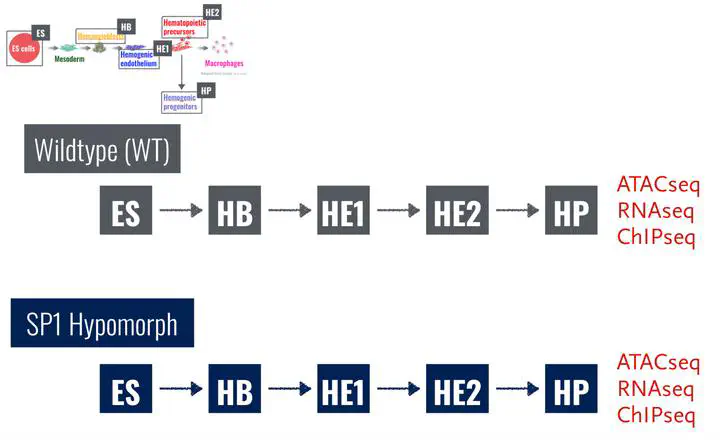
Dynamic alterations in gene expression programs during cell differentiation are regulated by the interplay of ubiquitous and tissue-specific transcription factors, and the epigenetic regulatory machinery. A mechanistic understanding can be postulated based on time-course multi-omics genome-wide measurements.
We developed a novel Bayesian nonparametric regression approach called Multi-Omics NETwork inference (MONET) for high-dimensionality multi-modal data integration to quantify the temporally dependent forces driving development and investigate by perturbation effects. Sparsity-inducing priors regularise the TF-gene interaction network and a Dirichlet process prior to regulate the number of participating TFs. This gives rise to a Bayesian nonparametric model where the number of TFs are learnt from data and not specified a priori. Statistical inference is performed in a Bayesian setting using an implementation of Hamiltonian Monte Carlo (HMC)-derived No-U Turn Sampler (NUTS) in STAN.
We show that MONET is able to extract accurate time-dependent TF-gene interaction networks and assess the importance of each TF for the different stages in haematopoietic development. Our model allows us to make predictions on the importance of specific TF over time and under different perturbation settings. Based on this we can create hypotheses for intervention to improve efficiency or halt development.
Researchers: